MICROPROTEIN-BASED TOOLS: ENABLING INHIBITION OF CHALLENGING DRUG TARGETS IN METASTATIC CANCER
In the last few decades of drug development, there have been limited improvements in cancer-related deaths per 100,000 people for most cancer types (Figure 1). This is largely due to the lack of curative treatment options for aggressive and treatment-resistant cancers with systemic spreading.
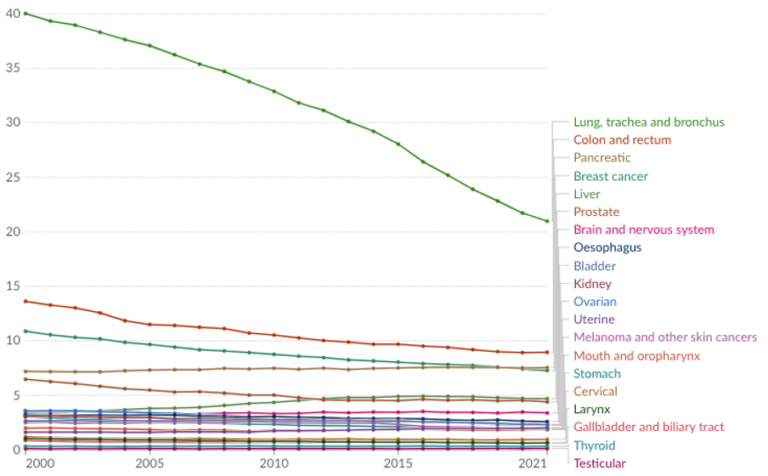
Figure 1. Cancer death rates by type, United States, the past 25 years. The reported annual death rate from different types of cancer per 100,000 people, based on the underlying cause listed on death certificates. Source: WHO Mortality Database (2024). Note: To allow for comparisons between countries and over time, this metric is age-standardized.
Treatment resistance and metastasis are driven by a class of very challenging drug targets (transcription factors) where conventional drug-development modalities have proven largely unsuccessful in producing inhibitors with clinical potential (Figure 2). The unstructured three-dimensional form and complex interaction network in the cells are important factors that make these factors so problematic to target. Traditional drug developmental screens and current treatment approaches are unable to regulate these factors efficiently. As such, there is a large unmet need to develop new novel strategies to identify and refine inhibitors toward challenging targets driving treatment resistance and systemic spreading to enable us to reduce cancer mortality.
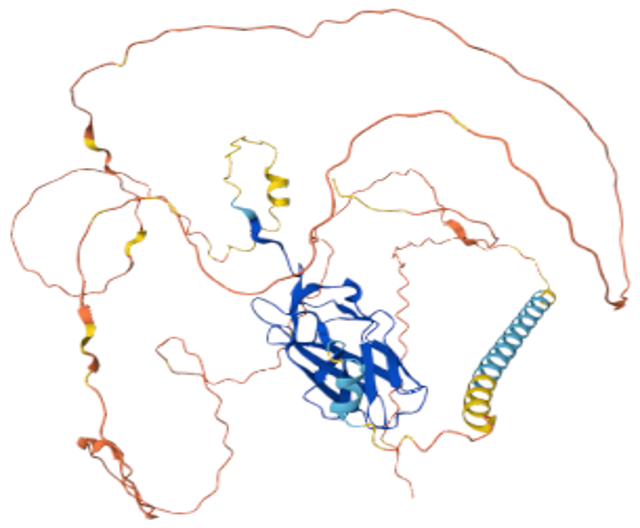
Figure 2. Unstructured transcription factor (RUNX2). Predicted three-dimensional structure of a representative transcription factor, RUNX2, showcasing a typically well-structured DNA binding domain (dark blue, central beta-sheets), otherwise a highly unstructured and flexible protein structure (“red thread” with only minor defined alpha-helical and beta-sheet structured regions). These unstructured regions provide advantageous complex flexibility for natural regulation in our cells but also pose a significant challenge in the quest to develop direct inhibitors toward these challenging targets.
MICROPROTEIN-BASED TOOLS HAVE TRANSFORMATIVE POTENTIAL FOR DEVELOPING DRUGS FOR INTRINSICALLY UNSTRUCTURED PROTEINS
Our novel microprotein approach provides a new strategy to identify and develop inhibitors of challenging cancer drivers. Microproteins naturally occur as small inhibitors and activators of larger proteins in our cells, which are currently blind spots in the genome discovery algorithms used (Figure 3). At the same time, a rapidly expanding field of research is demonstrating that these small proteins have essential roles in health and disease (see publications list for more information).
“Therefore, you can sequence the whole human genome and never know if a microprotein, like the recently identified ones, existed because it’s too short and falls below the usual length requirement for gene assignment algorithms”, Marie Rogne, CEO of Pioneer Research, explains. “This powerful reservoir of natural molecular regulators, if identified and generated into systematic tools, can harness the benefits of nature’s strategies to regulate the activity of difficult disease-driving proteins to develop new novel inhibitors for the clinic. The tools Pioneer Research are developing have great potential to identify natural inhibitors driving a wide array of diseases, including various cancers and neurological diseases and genetic syndromes”.
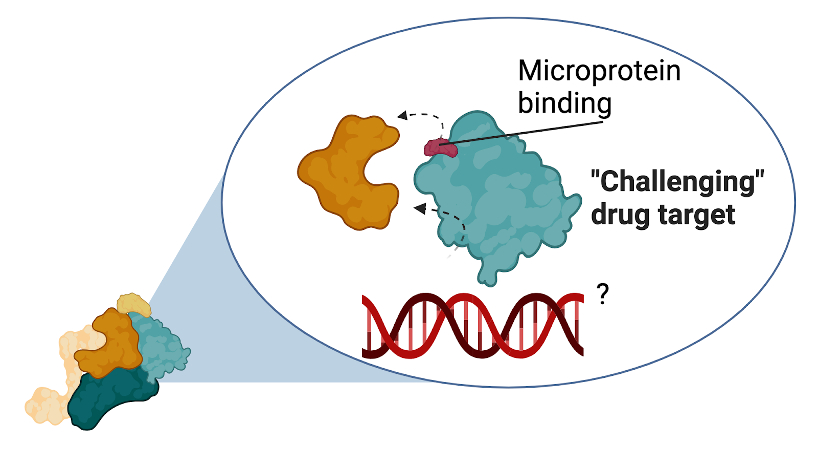
Figure 3. Microproteins bind to and regulate the activity of larger proteins. Recent discoveries have shown that microproteins can specifically bind and regulate the activity and/or affect the protein complex composition of its binding partners.
We have proof-of-concept evidence for the potent inhibition of two sought-after cancer-driving transcription factors under pre-clinical development. Showcasing the potential of our comprehensive, novel, and expression-validated microprotein screening platform generated to meet the need for systematic identification and development of inhibitors for challenging “unstructured” protein targets (Figure 4).
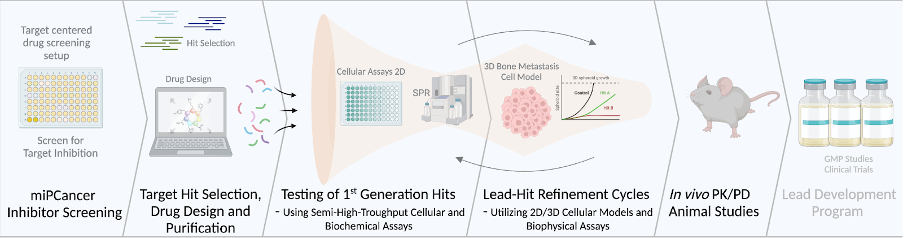
Figure 4. Workflow of the miPCancer Screening platforms. First, a target-centric activity-based cellular approach identifies microprotein inhibitors of challenging drug targets of interest. The inhibitor is refined through cycles of optimized activity assays and cellular assays to optimize, amongst others, biological effects, binding kinetics, and stability. Inhibitors with therapeutic potential are further examined in 3D cellular and animal models.
